Check out our literature!
Get a quick sense of what's OntoPortal with our short presenting paper . Please, use this to reference to our work too.
You can also discover OntoPortal related documents on our Zenodo Community and/or Zotero Collection .
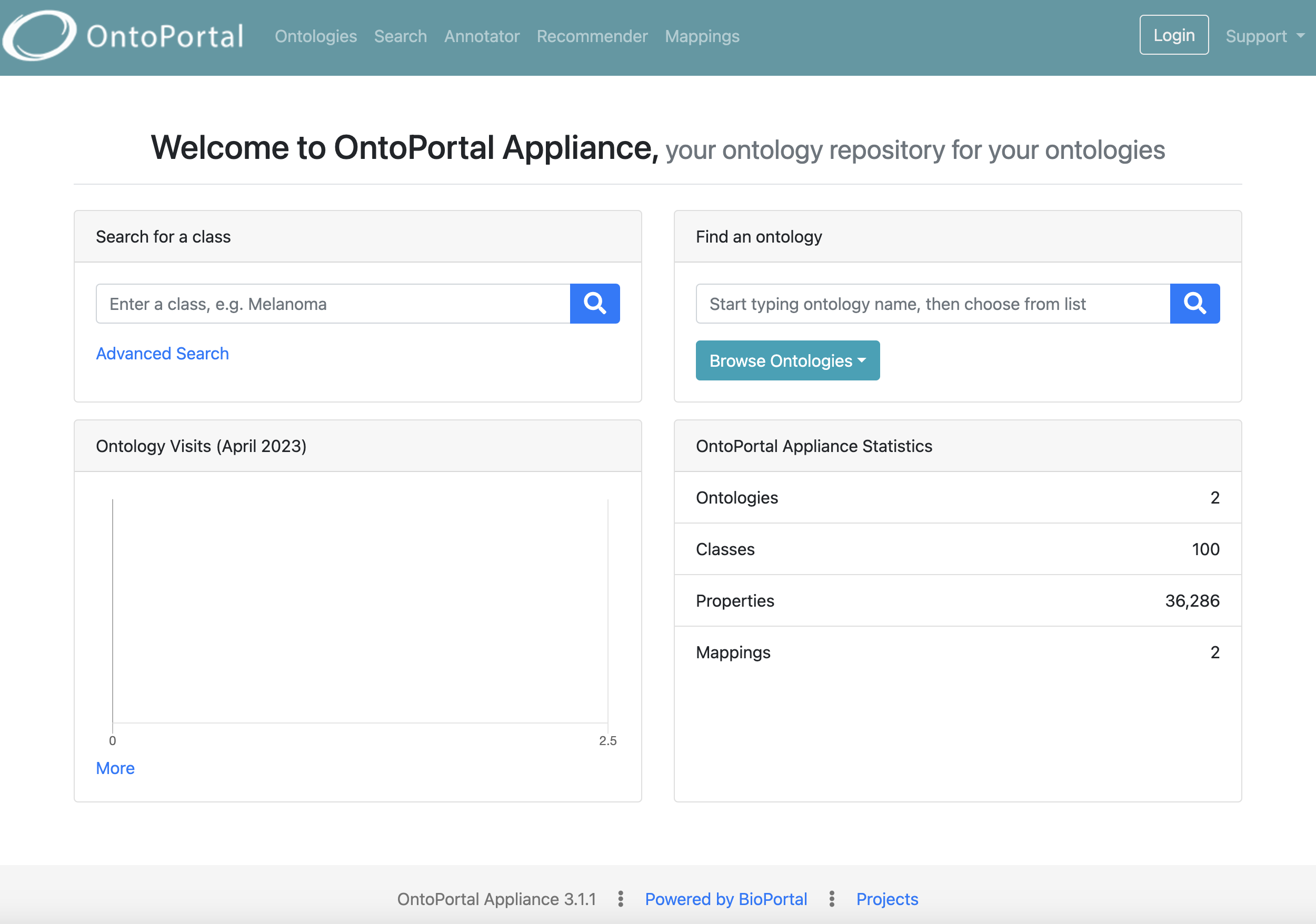
Save and find semantic artefacts
The primary mission of an OntoPortal installation is to host and serve ontologies and semantic artefacts. The portals accept resources in multiple knowledge representation languages: OWL, RDFS, SKOS, OBO and UMLS-RRF.
Ontologies are semantically described with rich metadata (partially extracted from the source files), and a browsing user interface allows to quickly identify, with faceted search, the ontologies of interest based on their metadata.
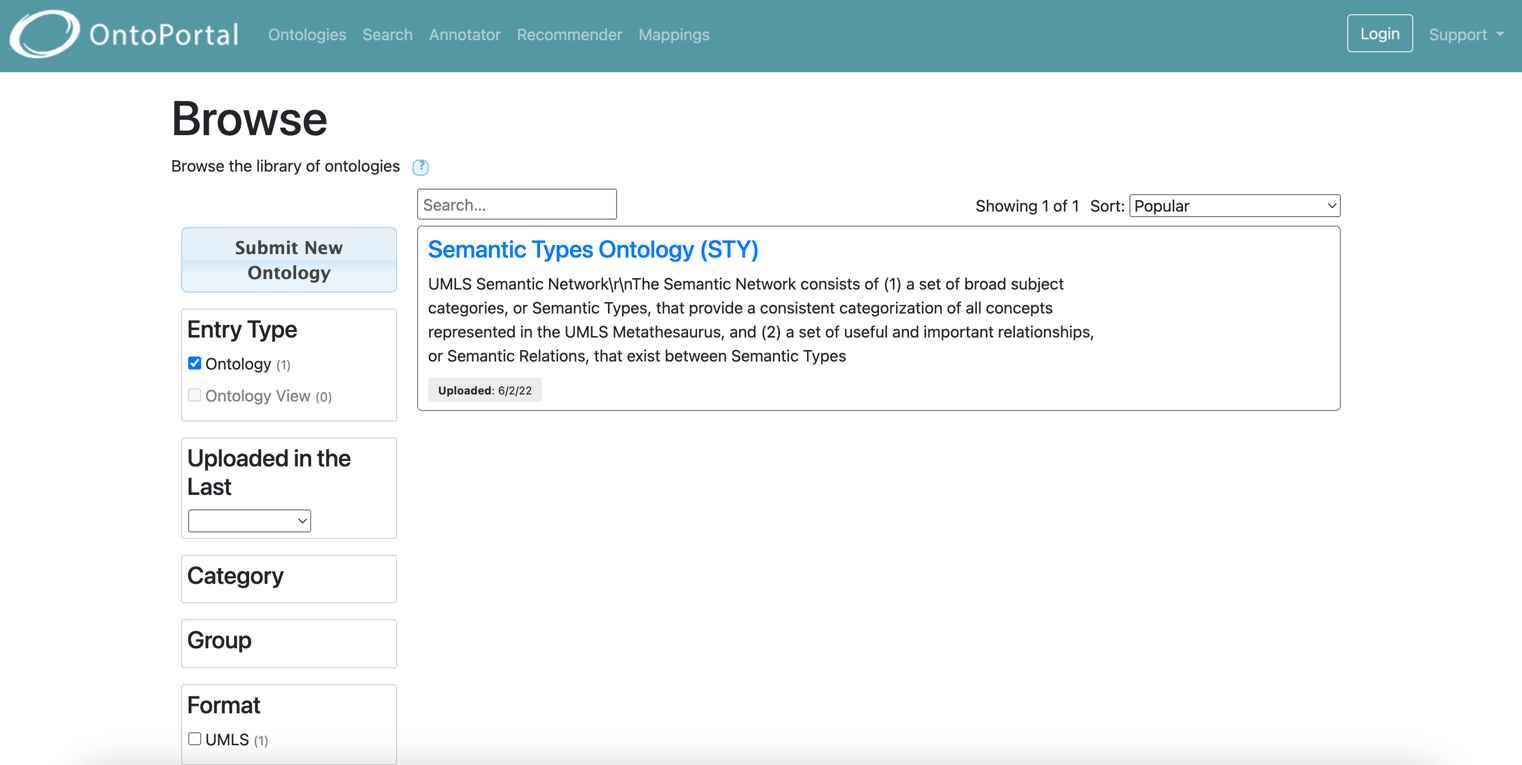
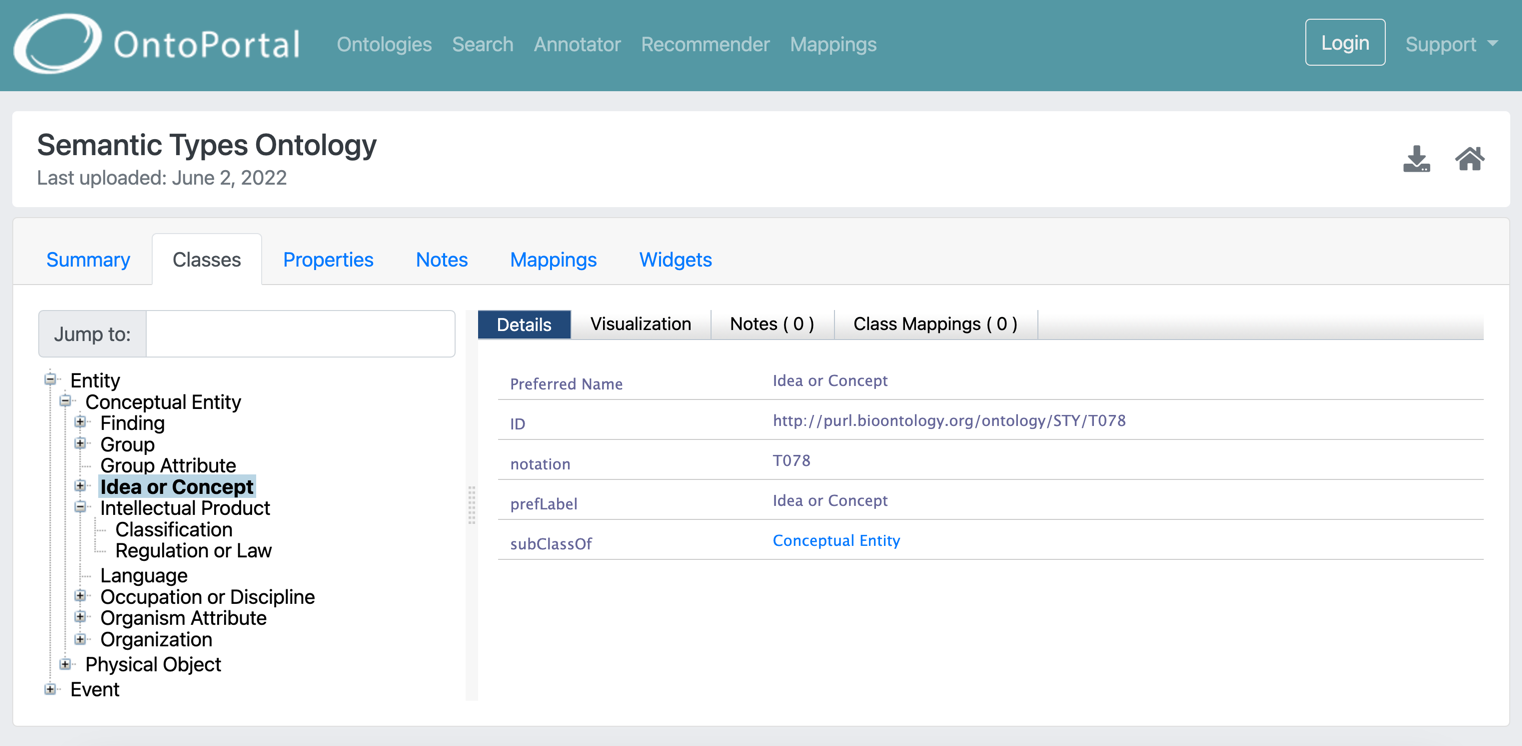
Browse the content of semantic artefacts
OntoPortal lets users visualize a class/concept or property within its hierarchy, as well as see related information for this entity (as relations included in the source file).
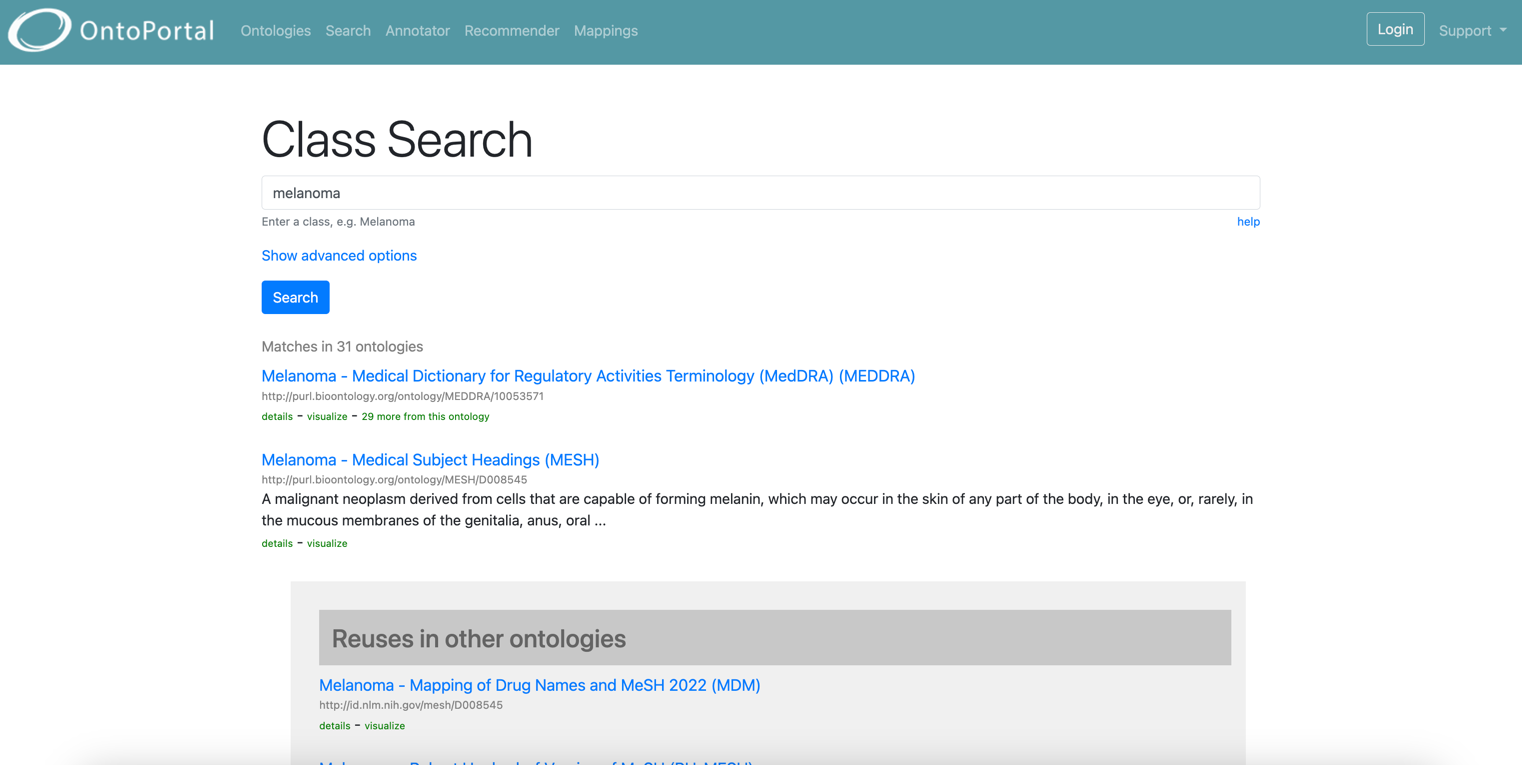
Search for terms
All the semantic artefacts content hosted in an OntoPortal is indexed and can easily be retrieved.
Much more available as standard OntoPortal services...
-
Custom hostingOrganize public/private ontologies, group/categorize them and offers custom slices.
-
VersioningStore all ontology versions, whether manually submitted or automatically pulled. Get diff between versions.
-
WidgetsEasily embed Web widgets in external web applications to facilitate the reuse/visualization of ontology entities.
-
MappingsStore, retrieve and explore mappings between ontology terms. Get lexical overlap of an ontology with all the other ones in a portal.
-
CommunityGet notified from feedback as notes, change requests and record projects using ontologies.
-
AnnotatorIdentify semantic artefact classes or concepts inside any raw text.
-
RecommenderGet ontology recommendations for a provided text or keyword list.
-
Automated accessAccess all content and services with two different endpoints: a REST web service API that returns JSON-LD and a SPARQL endpoint.
-
Standard ontology metadata modelStandardized and harmonized metadata model based on MOD1.4 to better support descriptions of ontologies and their relations.
-
Extended SKOS supportFull support of SKOS and innovative browsing approaches to discover and navigate concepts in SKOS thesauri that make extensive use of scheme and collections.
-
Multilingual supportFull support (search and browsing) of multilingual content within ontologies.
-
Generic Triple Store BackendStore and query ontology data through standards RDF triple store backend (4store, AllegroGraph, Virtuoso, GraphDB) for maximum scalability and interoperability.
And even more additional features and services developed by the Alliance...
-
Enhanced mapping featuresNew visualization and more flexible mapping storage, import mappings in bulk or extract SKOS mappings explicitly defined in semantic artefact source files.
-
Enhanced semantic annotation workflowNatural language-based enhancements of the Annotator with scoring methods context detection (e.g., negation, temporality).
-
Semantic artefact FAIRness assessmentGet FAIR score for each semantic artefact from 61 questions implemented from the Ontology FAIRness Evaluation (O’FAIRe) methodology.
-
Assigning DOIsRequest a Digital Object Identifier (DOI) for resources hosted in the portal using Datacite services.
-
New user interfacesEasy-to-use modern and respnsive user interfaces.
-
URIs dereferencingEasy-to-use infratructure supporting ontology URI dereferencing (resolutuon and content negociation).
-
Agents profilingSearch, browse, and manage agents (persons and organizations) associated to ontologies and see their complete profiles.
-
Related projectsRewamped ontology related project functionalities inclduing connectors to funded project databases.
-
MOD-API supportFull (level 4) MOD-API (v1) implementation for enhanced interoperability with other Semantic Artefact Catalogues.
-
FederationHarmonized search and discoveries of ontologies accross multiple federated OntoPortals.
-
Change requestsSuggest ontology content changes (new terms, synonyms, definitions) direclty from OntoPortal to GitHub.
-
Authentication & SSOIntegrate with LDAP or OAuth2 for unified access management.
What Users Say
Be part of a community
-
 The world's most comprehensive repository of biomedical ontologies
The world's most comprehensive repository of biomedical ontologies -
 A repository for French biomedical terminologies and ontologies
A repository for French biomedical terminologies and ontologies -
 A vocabulary and ontology repository for agronomy and related domains
A vocabulary and ontology repository for agronomy and related domains -
 The LifeWatch ERIC repository of semantic resources for ecology and related domains
The LifeWatch ERIC repository of semantic resources for ecology and related domains -
 A repository for Chinese biomedical terminologies and ontologies
A repository for Chinese biomedical terminologies and ontologies -
 The ontology repository for materials science
The ontology repository for materials science -
 A common ontology portal for industry and related domains
A common ontology portal for industry and related domains -
 A semantic artefact repository dedicated to Earth sciences
A semantic artefact repository dedicated to Earth sciences -
 A semantic artefact repository for biodiversity
A semantic artefact repository for biodiversity -
 A repository for the technology sciences domain
A repository for the technology sciences domain